Authors:
(1) Taha Laroussi, Laboratoire d’Hydrodynamique (LadHyX), CNRS, Ecole Polytechnique, Institut Polytechnique de Paris, 91120 Palaiseau, France;
(2) Mojtaba Jarrahi, Universit´e Paris-Saclay, CNRS, FAST, 91405 Orsay, France;
(3) Gabriel Amselem, Laboratoire d’Hydrodynamique (LadHyX), CNRS, Ecole Polytechnique, Institut Polytechnique de Paris, 91120 Palaiseau, France.
Table of Links
Discussion, Acknowledgements and References
V. DISCUSSION
We have shown experimentally that the change in phototactic behavior of C. reinhardtii can be due to its recent history of illumination. When exposed to two closely spaced, identical stimuli, the algae act as if they were time-integrating the stimuli. This is visible when the stimulus is close to that delimiting positive and negative phototaxis. Then, the algae show two different behaviors in response to the two consecutive stimuli: first a backand-forth response, then a negative phototaxis. When the stimuli are sufficiently spaced apart, the algae have had time to relax to a basal state, and show the same qualitative response to the two stimuli, see Fig. 6. In our experiments, we find this time of complete relaxation to be approximately equal to 30 minutes.
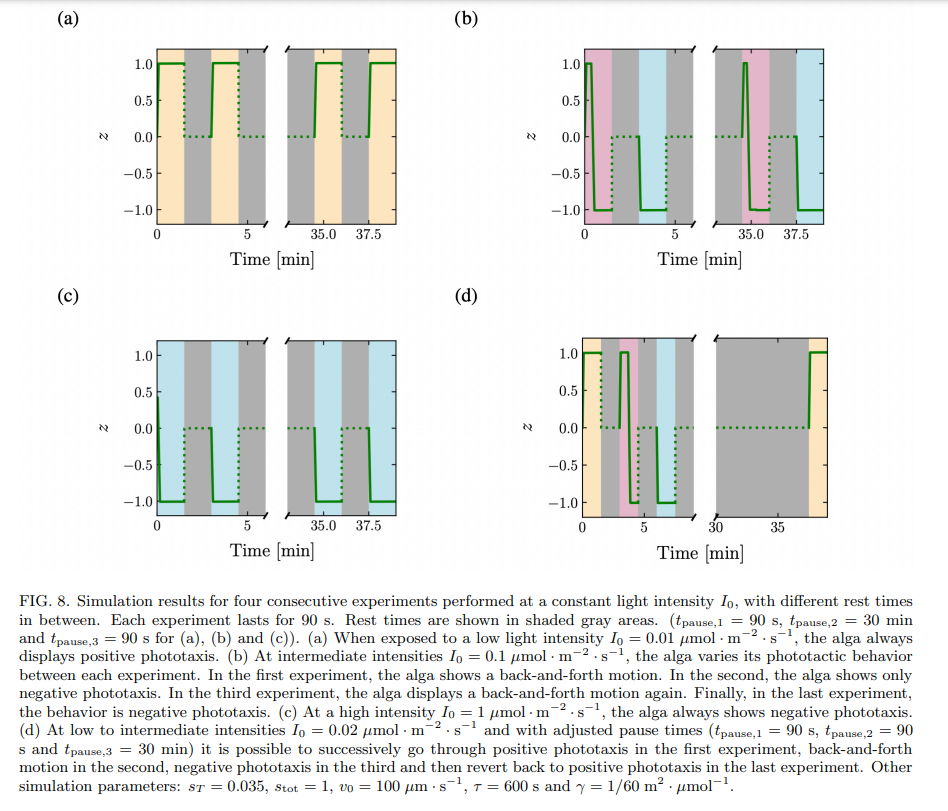
A simplified model of phototaxis explains this memory effect by the introduction of a time-scale of relaxation in the inner biochemistry of the cell. When the time-scale of relaxation is larger than the time between two consecutive stimuli, the alga integrates the signal and can display different behaviors in response to the two identical stimuli, see Fig. 8. The time needed to return to the basal state should be of order 3τ , which underlies our choice of τ = 10 min for the relaxation time constant. Our experimental data are consistent with an inner relaxation time τ in the range ≈ 5 to 20 minutes. What could be the biochemical species corresponding to the intracellular species S introduced in the model? It is very likely that our simplified model compounds the dynamics of multiple biochemical species, activated sequentially in a signalling cascade. Still, it is known that the photoreceptors for phototaxis in C. reinhardtii are channelrhodopsins, that become phosphorylated within milliseconds after the application of a light stimulus. A high level of phosphorylated channelrhodopsin-1 (ChR1) in the cell correlates with a change from positive to negative phototaxis [11], much like a high level of activated S corresponds to negative phototaxis in our model. Moreover, the time scale of dephosphorylation of ChR1 in C. reinhardtii has been shown to be of the order of ≈ 5 to 15 minutes [11], which is compatible with our value of τ = 10 min. A possibility is therefore to identify S with ChR1.
A more physical model of phototaxis was recently introduced by Leptos et al. to describe the response of C. reinhardtii to light [22]. In their model, Leptos et al. take into account the rotation of the microalga around its axis while swimming, and the resulting oscillating light intensity seen by the eyespot. The consecutive step-up and step-down stimuli sensed by the eyespot during one body rotation lead to a realignment of the trajectory towards the light source. Our model averages out the body rotation of the microalgae and describes the behavior of the microorganism at a more coarse-grained time-scale. Another difference is that the model of Leptos et al. does not take into account the fact that phototaxis changes sign when the light intensity increases. This change of phototactic behavior requires the introduction of a threshold value in the model, below (resp. above) which phototaxis is positive (resp. negative). Such a threshold is incorporated in our simplified model.
Note that the threshold in the model, introduced as a simple means to obtain a change in behavior between positive and negative phototaxis at different light intensities, leads to the back-and-forth behavior at intermediate light intensities. The existence of this threshold also enables to explain why the time scale of negative phototaxis is longer than that of positive phototaxis. Negative phototaxis occurs when the amount of active species s ⋆ overcomes a threshold sT ; this takes time, during which the alga exhibits positive phototaxis. Only when s ⋆ > sT do the algae show negative phototaxis, and accumulate at one boundary of the well. The small initial positive phototactic response predicted by the model is in agreement with experimental data. The model also predicts that it is possible to switch the phototactic behavior of algae from positive phototaxis to back-and-forth to negative phototaxis, using three consecutive identical stimuli, by adjusting the waiting times between the stimuli and choosing carefully the stimulus intensity, see Fig. 8d. While we were not able to find the experimental conditions corresponding to this theoretical prediction, we were able to experimentally observe the switch from back-and-forth to negative phototaxis, and from positive phototaxis to back-and-forth in a couple experiments, see Supp. Fig. 5.
While the model successfully reproduces the phototactic behavior and the time scales at play, it fails to reproduce two experimental observations. First, in a couple experiments, multiple consecutive back-and-forth of the center of mass were observed at intermediate intensities, see Supp. Fig. 6. Second, in experiments with consecutive stimuli at intermediate light intensities, algae in the third experiment (after a waiting time of 30 minutes) display positive phototaxis instead of the expected backand-forth behavior observed during the first experiment, see Fig. 6b and Supp. Fig. 5. Both behaviors can be obtained in the model by supposing that the threshold sT increases with the time spent under a light stimulus, and so depends on the history of the microorganism. Such an increasing threshold would also enable to recover the traditional adaptative behavior at longer time scales [15]. This implies having a third equation describing the timeevolution of sT , thus introducing another time constant into the problem.
In the lab, the memory effects reported here imply that it is not possible to repeat the same phototaxis experiments on a given batch of cells and expect similar outcomes. Such an effect was already known in the case of long-term adaptation [15]. The data reported here show that even in the case of short-term experiments, the behavior is not necessarily reproducible. To study phototaxis quantitatively, experimental repeats should be carried out with different batches of naive cells. Having identified this short time-scale, our experiments pave the way for the study of phototaxis in the presence of timevarying light stimuli, much as the algae are subject to in their natural environment. There, light is constantly fluctuating. The short-term memory could then be a beneficial way to integrate a light signal randomly interrupted by the shadow of other motile organisms or objects.
ACKNOWLEDGMENTS
We thank David Gonzalez-Rodriguez for insightful comments on an early version of the model, Maxime Simon for preliminary experiments, Salome Guttierez Ramos for help with microfabrication, and Caroline Frot for precious technical support. This work was supported by “Investissements d’Avenir” LabEx PALM (ANR-10- LABX-0039-PALM) and by PEPS “M´ecanique du futur”.
References
[1] G. J´ekely, Philosophical Transactions of the Royal Society B: Biological Sciences 364, 2795 (2009).
[2] K. W. Foster and R. D. Smyth, Microbiological reviews 44, 572 (1980).
[3] P. Hegemann and P. Berthold, in The Chlamydomonas Sourcebook (Elsevier, 2009) pp. 395–429.
[4] U. R¨uffer and W. Nultsch, Cell motility and the cytoskeleton 18, 269 (1991).
[5] M. E. H. Feinleib and G. M. Curry, Physiologia Plantarum 25, 346 (1971).
[6] W. Nultsch, J. Pfau, and R. Dolle, Archives of microbiology 144, 393 (1986).
[7] N. Morel-Laurens, Photochemistry and Photobiology 45, 119 (1987).
[8] R. Dolle, J. Pfau, and W. Nultsch, Journal of plant physiology 126, 467 (1987).
[9] T. Takahashi and M. Watanabe, FEBS Letters 336, 516 (1993).
[10] K.-i. Wakabayashi, Y. Misawa, S. Mochiji, and R. Kamiya, Proceedings of the National Academy of Sciences 108, 11280 (2011).
[11] M. B¨ohm, D. Boness, E. Fantisch, H. Erhard, J. Frauenholz, Z. Kowalzyk, N. Marcinkowski, S. Kateriya, P. Hegemann, and G. Kreimer, The Plant Cell 31, 886 (2019).
[12] R. M. Macnab and D. E. Koshland Jr, Proceedings of the National Academy of Sciences 69, 2509 (1972).
[13] H. C. Berg and P. Tedesco, Proceedings of the National Academy of Sciences 72, 3235 (1975).
[14] P. Van Haastert, The Journal of cell biology 96, 1559 (1983).
[15] A. Mayer, Nature 217, 875 (1968).
[16] E. Harris, D. Stern, and G. Witman, The chlamydomonas sourcebook, vol. 1 san diego (2009).
[17] Y. Xia and G. M. Whitesides, Annual review of materials science 28, 153 (1998).
[18] N. Ueki, T. Ide, S. Mochiji, Y. Kobayashi, R. Tokutsu, N. Ohnishi, K. Yamaguchi, S. Shigenobu, K. Tanaka, J. Minagawa, et al., Proceedings of the National Academy of Sciences 113, 5299 (2016).
[19] A. Ramamonjy, J. Dervaux, and P. Brunet, Physical Review Letters 128, 258101 (2022).
[20] C. T. Kreis, M. Le Blay, C. Linne, M. M. Makowski, and O. B¨aumchen, Nature Physics 14, 45 (2018).
[21] S. Till, F. Ebmeier, A. A. Fragkopoulos, M. G. Mazza, and O. B¨aumchen, Physical Review Research 4, L042046 (2022).
[22] K. C. Leptos, M. Chioccioli, S. Furlan, A. I. Pesci, and R. E. Goldstein, Physical Review E 107 (2023).
This paper is available on arxiv under CC 4.0 license.